Covariance derivative (wrt. x) under scaling (~ warping)
“A non-stationary covariance-based Kriging method for metamodelling in engineering design” Y. Xiong, W. Chen, D. Apley, and X. Ding, Int. J. Numer. Meth. Engng, 2007
if (!is.element("DiceKriging",installed.packages())) install.packages("DiceKriging")
# More robust (and expensive) EI optimization
if (!is.element("rgenoud",installed.packages())) install.packages("rgenoud")
# Also load DiceView for easy plot
if (!is.element("DiceView",installed.packages())) install.packages("DiceView")
# And DiceOptim for max_EI
if (!is.element("DiceOptim",installed.packages())) install.packages("DiceOptim")
library(DiceKriging)
packageDescription("DiceKriging")## Package: DiceKriging
## Title: Kriging Methods for Computer Experiments
## Version: 1.5.6
## Date: 2018-10-08
## Author: Olivier Roustant, David Ginsbourger, Yves Deville.
## Contributors: Clement Chevalier, Yann Richet.
## Maintainer: Olivier Roustant <roustant@emse.fr>
## Description: Estimation, validation and prediction of kriging
## models. Important functions : km, print.km, plot.km,
## predict.km.
## Depends: methods
## Suggests: rgenoud (>= 5.8-2.0), foreach, doParallel, testthat,
## numDeriv
## License: GPL-2 | GPL-3
## URL: https://dicekrigingclub.github.io/www/
## NeedsCompilation: yes
## Packaged: 2018-10-08 10:02:28 UTC; travis
## Repository: CRAN
## Date/Publication: 2018-10-08 10:50:13 UTC
## Built: R 3.5.1; x86_64-pc-linux-gnu; 2018-11-23 01:01:26 UTC;
## unix
##
## -- File: /home/travis/R/Library/DiceKriging/Meta/package.rds# library(testthat)Covariance derivative (wrt. x)
f = function(x) {
x = x^4
1-1/2*(sin(12*x)/(1+x)+2*cos(7*x)*x^5+0.7)}
plot(f)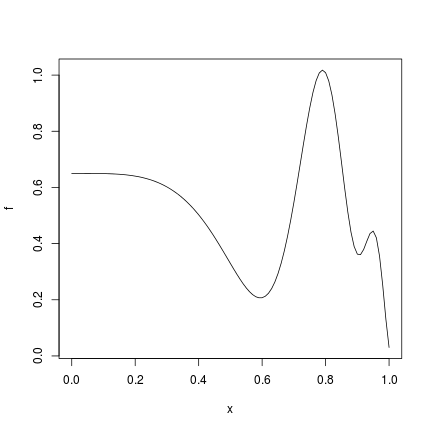
Checking km covariance derivative ‘covVector.dx’
X <- matrix(c(0,.25,.5,.75,1.0),ncol=1)
y <- f(X)
set.seed(123)
m <- km(formula=~1, design=X, response=y,scaling=F,control=list(trace=FALSE))
print(m)##
## Call:
## km(formula = ~1, design = X, response = y, control = list(trace = FALSE),
## scaling = F)
##
## Trend coeff.:
## Estimate
## (Intercept) 0.5039
##
## Covar. type : matern5_2
## Covar. coeff.:
## Estimate
## theta(design) 0.0000
##
## Variance estimate: 0.08705396# plot covariance function (of x)
x = seq(0,1,,101) #101 because we need to have X values in x to check derivative is null
cx = matrix(NaN,nrow=nrow(X),ncol=length(x)) # covariance function (in 5d space)
cdx = matrix(NaN,nrow=nrow(X),ncol=length(x)) # derivative of covariance function
for (i in 1:length(x)) {
mx = predict(m,x[i],type="SK",se.compute=T)
cx[,i] = covMatrix(m@covariance,rbind(X=m@X,x[i]))$C[1:length(X),1+length(X)]
cdx[,i] = covVector.dx(m@covariance,x=x[i],X=m@X,mx$c)
}
par(mfrow=c(2,3))
for (j in 1:length(X)) {
plot(x, cx[j,], type='l',main=paste0('Covariance to X_',j))
abline(v=X[j,],col='blue')
for (i in 1:(length(x)/10))
arrows(x[10*i], cx[j,10*i], x[10*i]+0.1, cx[j,10*i] + cdx[j,10*i]/10,length=0.05,col='red')
ij = which(x==X[j,1])
# testthat::test_that(desc="zero derivative at X_i",expect_true(cdx[j,ij] == 0))
}
par(mfrow=c(1,1))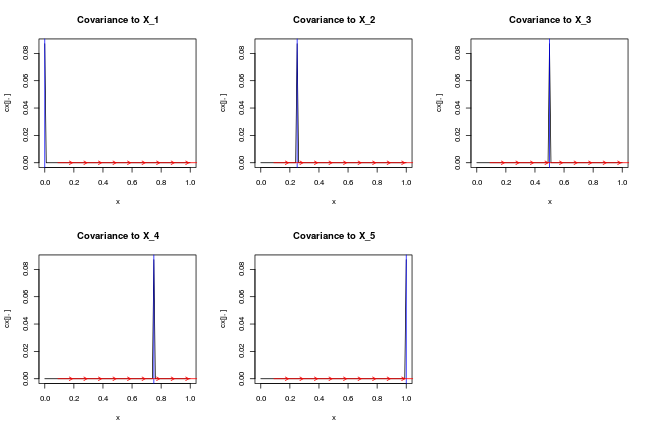
Checking km scaling (1 node) covariance derivative ‘covVector.dx’
X <- matrix(c(0,.25,.5,.75,1.0),ncol=1)
y <- f(X)
set.seed(123)
m_scaling0 <- km(formula=~1, design=X, response=y,scaling=T,knots=list(design=c(.5)),control=list(trace=FALSE))
print(m_scaling0)##
## Call:
## km(formula = ~1, design = X, response = y, control = list(trace = FALSE),
## scaling = T, knots = list(design = c(0.5)))
##
## Trend coeff.:
## Estimate
## (Intercept) 0.5019
##
## Covar. type : matern5_2 , with scaling
## Covar. coeff., with estimated values for eta:
##
## eta(design) 11.8879
## knots(design) 0.5000
##
## Variance estimate: 0.09009309# plot covariance function (of x)
x = seq(0,1,,101) #101 because we need to have X values in x to check derivative is null
cx = matrix(NaN,nrow=nrow(X),ncol=length(x)) # covariance function (in 5d space)
cdx = matrix(NaN,nrow=nrow(X),ncol=length(x)) # derivative of covariance function
for (i in 1:length(x)) {
mx = predict(m_scaling0,x[i],type="SK",se.compute=T)
cx[,i] = covMatrix(m_scaling0@covariance,rbind(X=m_scaling0@X,x[i]))$C[1:length(X),1+length(X)]
cdx[,i] = covVector.dx(m_scaling0@covariance,x=x[i],X=m_scaling0@X,mx$c)
}
par(mfrow=c(2,3))
for (j in 1:length(X)) {
plot(x, cx[j,], type='l',main=paste0('Covariance to X_',j))
abline(v=X[j,],col='blue')
for (i in 1:(length(x)/10))
arrows(x[10*i], cx[j,10*i], x[10*i]+0.1, cx[j,10*i] + cdx[j,10*i]/10,length=0.05,col='red')
ij = which(x==X[j,1])
# test_that(desc="zero derivative at X_i",expect_true(cdx[j,ij] == 0))
}
par(mfrow=c(1,1))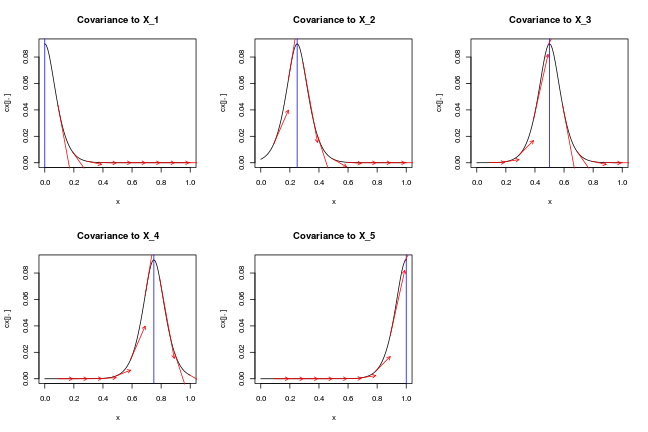
Checking km scaling covariance derivative ‘covVector.dx’
X <- matrix(c(0,0.125,.25,0.375,.5,0.625,.75,0.875,1.0),ncol=1)
y <- f(X)
set.seed(123)
m_scaling <- km(formula=~1, design=X, response=y,scaling=T,knots=list(design=c(0,.5,1)),control=list(trace=FALSE))
print(m_scaling)##
## Call:
## km(formula = ~1, design = X, response = y, control = list(trace = FALSE),
## scaling = T, knots = list(design = c(0, 0.5, 1)))
##
## Trend coeff.:
## Estimate
## (Intercept) 0.4395
##
## Covar. type : matern5_2 , with scaling
## Covar. coeff., with estimated values for eta:
##
## eta(design) 0.5000 2.6074 35.5075
## knots(design) 0.0000 0.5000 1.0000
##
## Variance estimate: 0.08215635# plot covariance function (of x)
x = seq(0,1,,201) #101 because we need to have X values in x to check derivative is null
cx = matrix(NaN,nrow=nrow(X),ncol=length(x)) # covariance function (in 5d space)
cdx = matrix(NaN,nrow=nrow(X),ncol=length(x)) # derivative of covariance function
for (i in 1:length(x)) {
mx = predict(m_scaling,x[i],type="SK",se.compute=T)
cx[,i] = covMatrix(m_scaling@covariance,rbind(X=m_scaling@X,x[i]))$C[1:length(X),1+length(X)]
cdx[,i] = covVector.dx(m_scaling@covariance,x=x[i],X=m_scaling@X,mx$c)
}
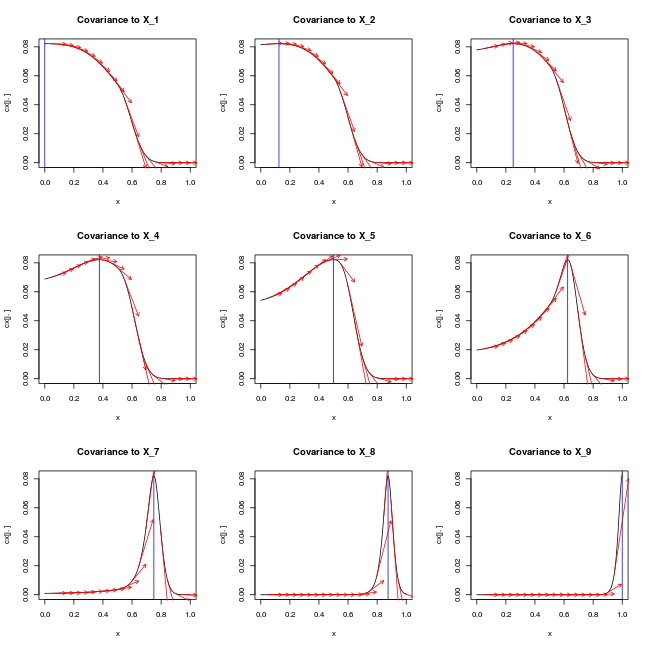
par(mfrow=c(3,3))
for (j in 1:length(X)) {
plot(x, cx[j,], type='l',main=paste0('Covariance to X_',j))
abline(v=X[j,],col='blue')
for (i in 1:(length(x)/10))
arrows(x[10*i], cx[j,10*i], x[10*i]+0.1, cx[j,10*i] + cdx[j,10*i]/10,length=0.05,col='red')
ij = which(x==X[j,1])
# test_that(desc="zero derivative at X_i",expect_true(cdx[j,ij] == 0))
}
par(mfrow=c(1,1))Checking km affine scaling (no node given) covariance derivative ‘covVector.dx’
X <- matrix(c(0,0.125,.25,0.375,.5,0.625,.75,0.875,1.0),ncol=1)
y <- f(X)
set.seed(123)
m_ascaling <- km(formula=~1, design=X, response=y,scaling=T,knots=NULL,control=list(trace=FALSE))
print(m_ascaling)##
## Call:
## km(formula = ~1, design = X, response = y, control = list(trace = FALSE),
## scaling = T, knots = NULL)
##
## Trend coeff.:
## Estimate
## (Intercept) 0.4293
##
## Covar. type : matern5_2 , with scaling
## Covar. coeff., with estimated values for eta:
##
## eta(design) 0.5000 11.1254
## knots(design) 0.0000 1.0000
##
## Variance estimate: 0.08701197# plot covariance function (of x)
x = seq(0,1,,201) #101 because we need to have X values in x to check derivative is null
cx = matrix(NaN,nrow=nrow(X),ncol=length(x)) # covariance function (in 5d space)
cdx = matrix(NaN,nrow=nrow(X),ncol=length(x)) # derivative of covariance function
for (i in 1:length(x)) {
mx = predict(m_ascaling,x[i],type="SK",se.compute=T)
cx[,i] = covMatrix(m_ascaling@covariance,rbind(X=m_ascaling@X,x[i]))$C[1:length(X),1+length(X)]
cdx[,i] = covVector.dx(m_ascaling@covariance,x=x[i],X=m_ascaling@X,mx$c)
}
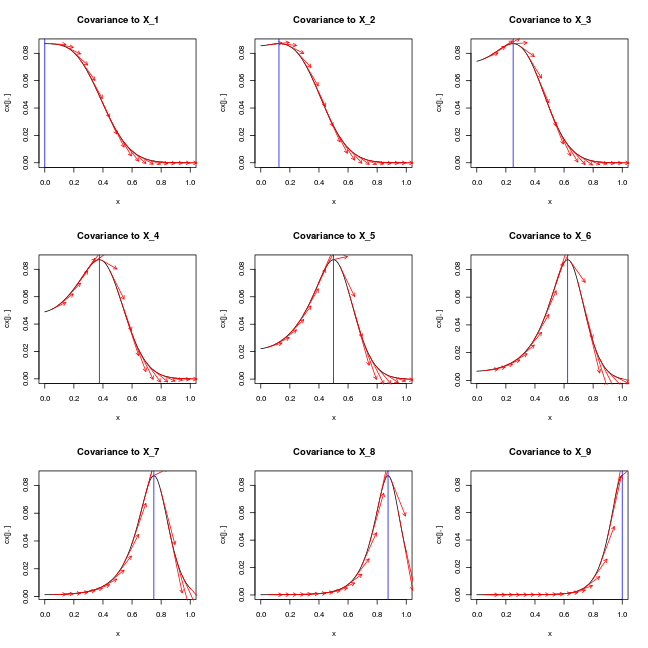
par(mfrow=c(3,3))
for (j in 1:length(X)) {
plot(x, cx[j,], type='l',main=paste0('Covariance to X_',j))
abline(v=X[j,],col='blue')
for (i in 1:(length(x)/10))
arrows(x[10*i], cx[j,10*i], x[10*i]+0.1, cx[j,10*i] + cdx[j,10*i]/10,length=0.05,col='red')
ij = which(x==X[j,1])
# test_that(desc="zero derivative at X_i",expect_true(cdx[j,ij] == 0))
}
par(mfrow=c(1,1))… used with EGO
## Package: DiceOptim
## Version: 2.0
## Title: Kriging-Based Optimization for Computer Experiments
## Date: 2016-09-06
## Author: V. Picheny, D. Ginsbourger, O. Roustant, with
## contributions by M. Binois, C. Chevalier, S. Marmin,
## and T. Wagner
## Maintainer: V. Picheny <victor.picheny@toulouse.inra.fr>
## Description: Efficient Global Optimization (EGO) algorithm and
## adaptations for parallel infill (multipoint EI),
## problems with noise, and problems with constraints.
## Depends: DiceKriging (>= 1.2), methods
## Imports: randtoolbox, pbivnorm, rgenoud, mnormt, DiceDesign
## Suggests: KrigInv, GPareto, lhs
## License: GPL-2 | GPL-3
## URL: http://dice.emse.fr/
## RoxygenNote: 5.0.1
## NeedsCompilation: yes
## Packaged: 2016-09-13 14:38:07 UTC; vpicheny
## Repository: CRAN
## Date/Publication: 2016-09-15 17:33:46
## Built: R 3.5.1; x86_64-pc-linux-gnu; 2018-11-23 08:16:13 UTC;
## unix
##
## -- File: /home/travis/R/Library/DiceOptim/Meta/package.rdsEGO.nsteps = function (model, fun, nsteps, lower, upper, parinit = NULL,
control = NULL, kmcontrol = NULL)
{
n <- nrow(model@X)
if (is.null(kmcontrol$penalty))
kmcontrol$penalty <- model@penalty
if (length(model@penalty == 0))
kmcontrol$penalty <- NULL
if (is.null(kmcontrol$optim.method))
kmcontrol$optim.method <- model@optim.method
if (is.null(kmcontrol$parinit))
kmcontrol$parinit <- model@parinit
if (is.null(kmcontrol$control))
kmcontrol$control <- model@control
control$print.level = 0
for (i in 1:nsteps) {
oEGO <- max_EI(model = model, lower = lower, upper = upper,
parinit = parinit, control = control)
model@X <- rbind(model@X, oEGO$par)
model@y <- rbind(model@y, fun(t(oEGO$par)))
kmcontrol$parinit <- covparam2vect(model@covariance)
kmcontrol$control$trace = FALSE
kmcontrol$scaling = is.element("eta",slotNames(model@covariance))
if (is.element("knots",slotNames(model@covariance))) kmcontrol$knots = model@covariance@knots else kmcontrol$knots = NULL
if (model@param.estim) {
model <- km(formula = model@trend.formula, design = model@X,
response = model@y, covtype = model@covariance@name,
lower = model@lower, upper = model@upper, nugget = NULL,
penalty = kmcontrol$penalty, optim.method = kmcontrol$optim.method,
parinit = kmcontrol$parinit, control = kmcontrol$control,
gr = model@gr, iso = is(model@covariance, "covIso"),
scaling = kmcontrol$scaling, knots = kmcontrol$knots)
}
else {
coef.cov <- covparam2vect(model@covariance)
model <- km(formula = model@trend.formula, design = model@X,
response = model@y, covtype = model@covariance@name,
coef.trend = model@trend.coef, coef.cov = coef.cov,
coef.var = model@covariance@sd2, nugget = NULL,
iso = is(model@covariance, "covIso"))
}
}
return(list(par = model@X[(n + 1):(n + nsteps), , drop = FALSE],
value = model@y[(n + 1):(n + nsteps), , drop = FALSE],
npoints = 1, nsteps = nsteps, lastmodel = model))
}Objective function
branin = function(x) {
x = x^.5
DiceKriging::branin(x)
}
# a 9-points factorial design, and the corresponding response
d <- 2
n <- 9
design.fact <- expand.grid(seq(0,1,length=3), seq(0,1,length=3))
names(design.fact)<-c("x1", "x2")
design.fact <- data.frame(design.fact)
names(design.fact)<-c("x1", "x2")
response.branin <- apply(design.fact, 1, branin)
response.branin <- data.frame(response.branin)
names(response.branin) <- "y"
.x = seq(0,1,,51)
.p3d = persp(.x,.x,matrix(apply(expand.grid(.x,.x),1,branin),ncol=length(.x)),zlab = "branin(sqrt(.))", phi = 60,theta = 30)
points(trans3d(design.fact[,1],design.fact[,2],response.branin$y,.p3d),col='black',pch=20) 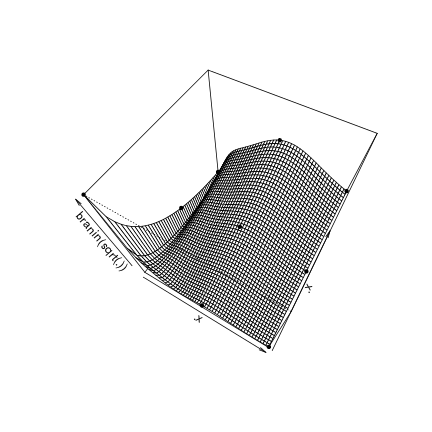
Basic (no scaling)
set.seed(123)
# model identification
fitted.model1 <- km(~1, design=design.fact, response=response.branin,
covtype="gauss", control=list(pop.size=50,trace=FALSE), parinit=c(0.5, 0.5))
# EGO n steps
library(rgenoud)## ## rgenoud (Version 5.8-2.0, Build Date: 2018-04-03)
## ## See http://sekhon.berkeley.edu/rgenoud for additional documentation.
## ## Please cite software as:
## ## Walter Mebane, Jr. and Jasjeet S. Sekhon. 2011.
## ## ``Genetic Optimization Using Derivatives: The rgenoud package for R.''
## ## Journal of Statistical Software, 42(11): 1-26.
## ##nsteps <- 10
lower <- rep(0,d)
upper <- rep(1,d)
oEGO <- EGO.nsteps(model=fitted.model1, fun=branin, nsteps=nsteps,
lower=lower, upper=upper, control=list(pop.size=20, BFGSburnin=2))
print(oEGO$par)## x1 x2
## [1,] 0.2875775 0.78830514
## [2,] 0.9404673 0.04555650
## [3,] 0.5514350 0.45661474
## [4,] 0.6775706 0.57263340
## [5,] 0.2460877 0.04205953
## [6,] 0.8895393 0.69280341
## [7,] 0.6557058 0.70853047
## [8,] 0.2891597 0.14711365
## [9,] 0.6907053 0.79546742
## [10,] 0.7584595 0.21640794print(oEGO$value)## y
## [1,] 121.203156
## [2,] 1.196258
## [3,] 102.920296
## [4,] 118.853490
## [5,] 2.701754
## [6,] 109.889793
## [7,] 149.223575
## [8,] 12.275818
## [9,] 162.495973
## [10,] 39.101408.p3d = persp(.x,.x,matrix(apply(expand.grid(.x,.x),1,branin),ncol=length(.x)),zlab = "branin(sqrt(.))", phi = 60,theta = 30)
points(trans3d(oEGO$lastmodel@X[,1],oEGO$lastmodel@X[,2],apply(oEGO$lastmodel@X,1,branin),.p3d),col='black',pch=20)
points(trans3d(oEGO$par[,1],oEGO$par[,2],apply(oEGO$par,1,branin),.p3d),col='red',pch=20)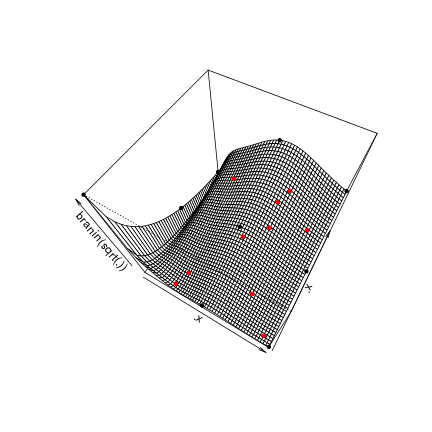
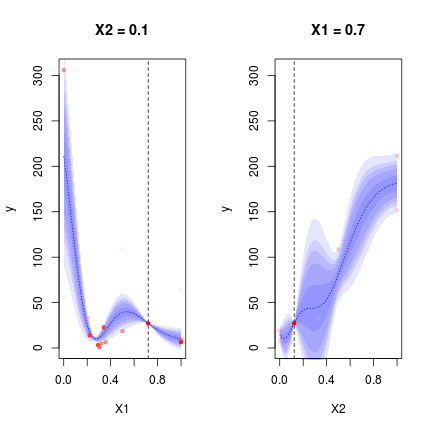
DiceView::sectionview.km(oEGO$lastmodel,center=oEGO$par[1,])## Warning in rgl.init(initValue, onlyNULL): RGL: unable to open X11
## display## Warning: 'rgl_init' failed, running with rgl.useNULL = TRUE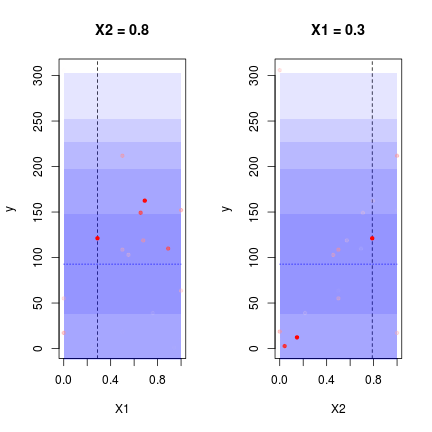
With scaling
set.seed(123)
# model identification
fitted.model1 <- km(~1, design=design.fact, response=response.branin,
covtype="gauss", control=list(pop.size=50,trace=FALSE), parinit=c(0.5, 0.5), scaling = T)
# EGO n steps
library(rgenoud)
nsteps <- 10
lower <- rep(0,d)
upper <- rep(1,d)
oEGO <- EGO.nsteps(model=fitted.model1, fun=branin, nsteps=nsteps,
lower=lower, upper=upper, control=list(pop.size=20, BFGSburnin=2))
print(oEGO$par)## x1 x2
## [1,] 0.71988164 0.12314325
## [2,] 0.09945093 0.72900214
## [3,] 0.34056115 0.17895932
## [4,] 0.35818519 0.00000000
## [5,] 0.32261555 0.00000000
## [6,] 1.00000000 0.10628820
## [7,] 0.22051423 0.17010558
## [8,] 0.30640514 0.02504285
## [9,] 0.29139243 0.06889040
## [10,] 0.20311376 0.32754022print(oEGO$value)## y
## [1,] 27.2155487
## [2,] 59.8182973
## [3,] 22.7085006
## [4,] 6.3605743
## [5,] 4.9462304
## [6,] 6.5254250
## [7,] 14.0956776
## [8,] 0.5847939
## [9,] 3.1345166
## [10,] 33.1293612.p3d = persp(.x,.x,matrix(apply(expand.grid(.x,.x),1,branin),ncol=length(.x)),zlab = "branin(sqrt(.))", phi = 60,theta = 30)
points(trans3d(oEGO$lastmodel@X[,1],oEGO$lastmodel@X[,2],apply(oEGO$lastmodel@X,1,branin),.p3d),col='black',pch=20)
points(trans3d(oEGO$par[,1],oEGO$par[,2],apply(oEGO$par,1,branin),.p3d),col='red',pch=20) 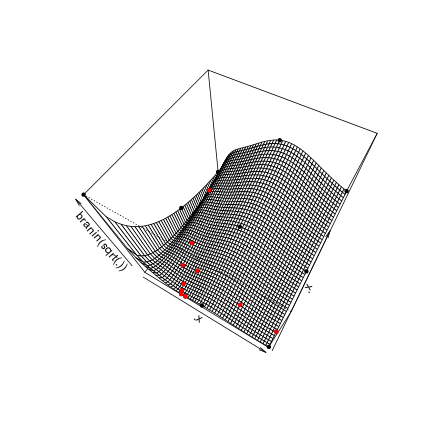
DiceView::sectionview.km(oEGO$lastmodel,center=oEGO$par[1,])